 A generalized PCR-based genetic targeting primer designer
A generalized PCR-based genetic targeting primer designer
Polymerase chain reaction (PCR) based gene deletion and tagging is a simple and powerful approach for studying gene functions. Although it was originally developed in classic yeast models such as Saccharomyces cerevisiae and Schizosaccharomyces pombe, the same principle equally applies to filamentous fungi and other eukaryotic systems with efficient homologous recombination. Moreover, rather than relying on a single reference strain, more and more studies nowadays have begun to explore higher levels of functional diversity and complexity using natural isolates, imposing a real need for functional genomics tools that are amenable for such use scenarios. Here we present GetPrimers, a generalized computational framework and web tool for automatically designing primers of PCR-based gene deletion and tagging. Two gene targeting methods are supported: the one-step PCR based method (the "long primer" mode of GetPrimers) and the fusion PCR based method (the "short primer" mode of GetPrimers). The one-step PCR based strategy is more conventionally used but the sizes of the homoglous regions that it creates for recombination-mediated gene targeting are usually limited (e.g. ~ 40 bp). In contrast, the fusion PCR based strategy can create homologous regions that are orders of magnitude larger and therefore substantially increases the efficency of recombination-mediated gene targeting. While pre-computed genome-wide results are provided for Saccharomyces cerevisiae and Schizosaccharomyces pombe reference genomes out of the box, the input genome, gene annotation, and plasmid sequences can be fully customized based on any given species and genetic backgrounds.
Note: With GetPrimers, we have shipped a number of most commonly used genomes and plasmids. In case this dose not suffice your needs, custome genome and plasmid sequences can always be uploaded and processed by GetPrimers. And if you believe that your custom genome or plasmid could be useful for many others in the field, do not hesitate to contact us (yuejiaxing[at]gmail[dot]com) to add your custome sequences to the pre-build database of GetPrimers.
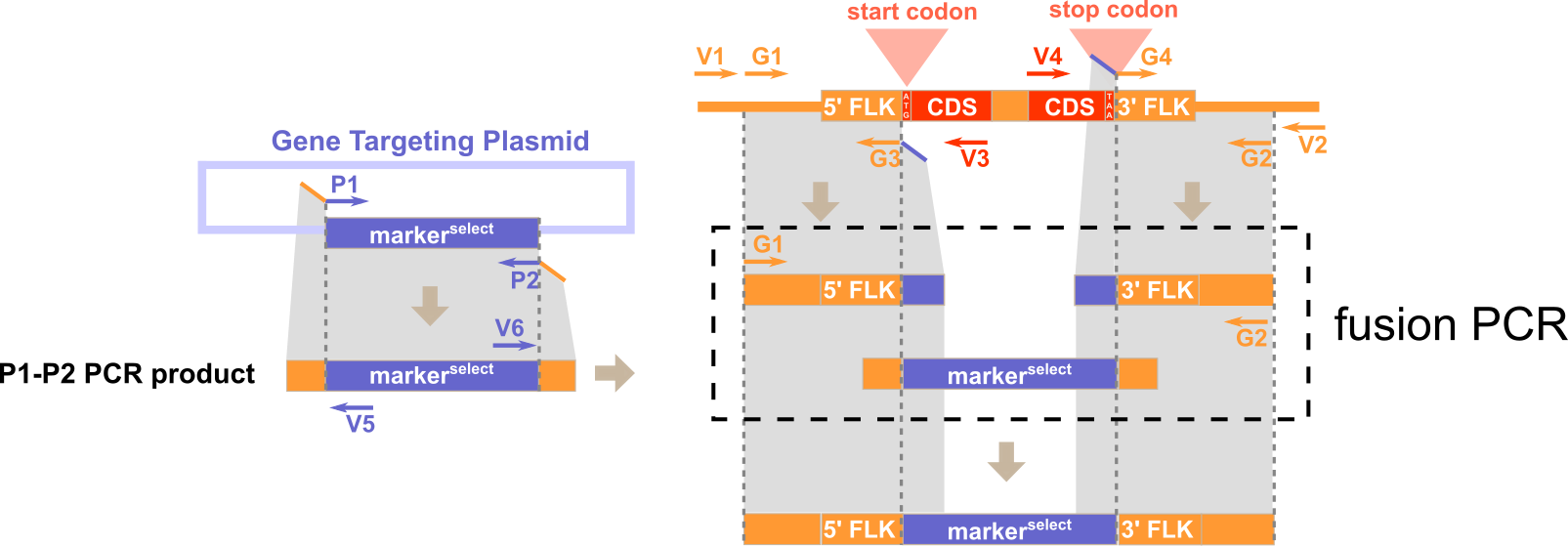
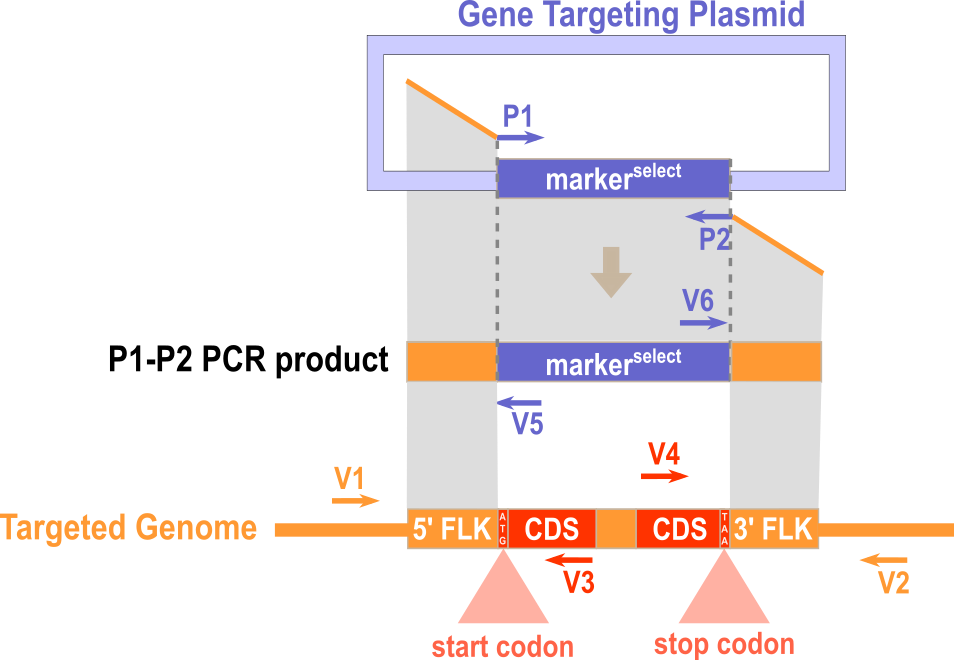
1) P1 and P2 for amplifying the inserted sequences (e.g. selection cassette) from the plasmid. 2) G1 and G3 for amplifying the 5'-flanking of the targeted genomic region. 3) G4 and G2 for amplifying the 3'-flanking of the targeted genomic region.
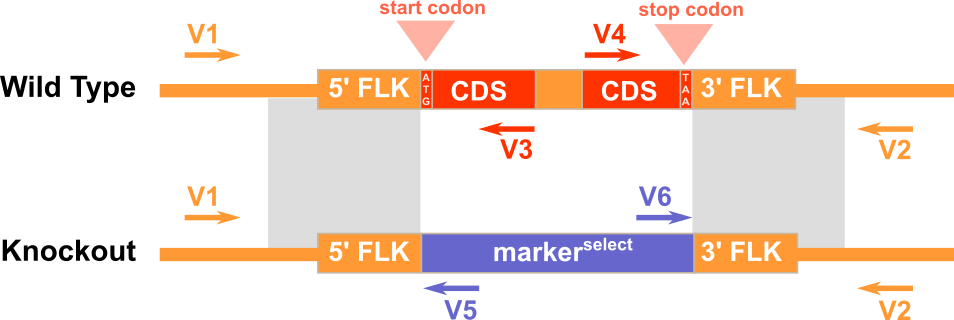
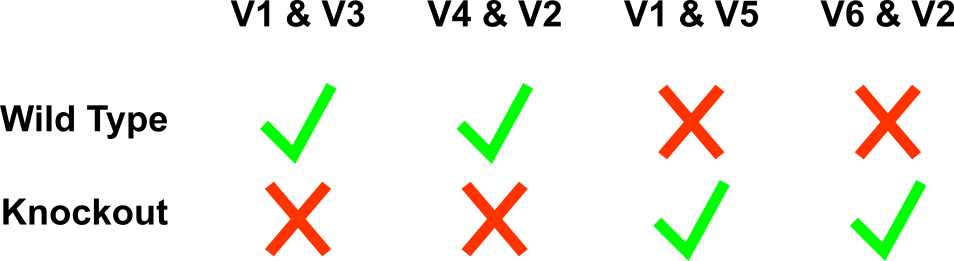
1) V1 and V3 for amplifying the 5'-junction of the WT genomic region before gene targeting. 2) V4 and V2 for amplifying the 3'-junction of the WT genomic region before gene targeting. 3) V1 and V5 for amplifying the 5'-junction the recombinant genomic region after gene targeting. 4) V6 and V2 for amplifying the 3'-junction the recombinant genomic region after gene targeting.
Experimental Design
Expected PCR results with verification primers
Note: G3 is located immediately upstream of the start codon and G4 is located immediately downstream of the stop codon.
Experimental Design
Expected PCR results with verification primers
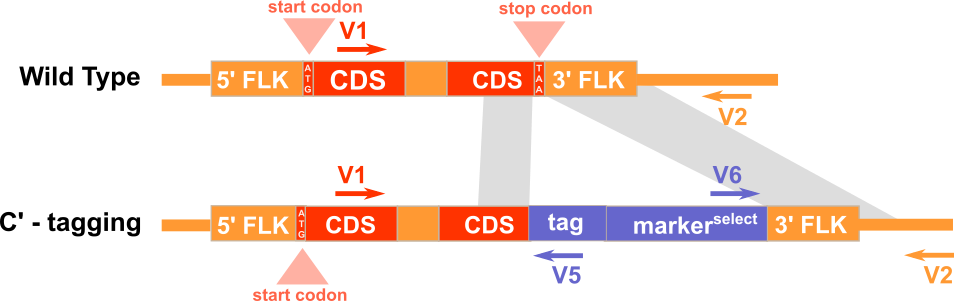

Note: G3 is located immediately upstream of the stop codon and G4 is located immediately downstream of the stop codon.
Experimental Design
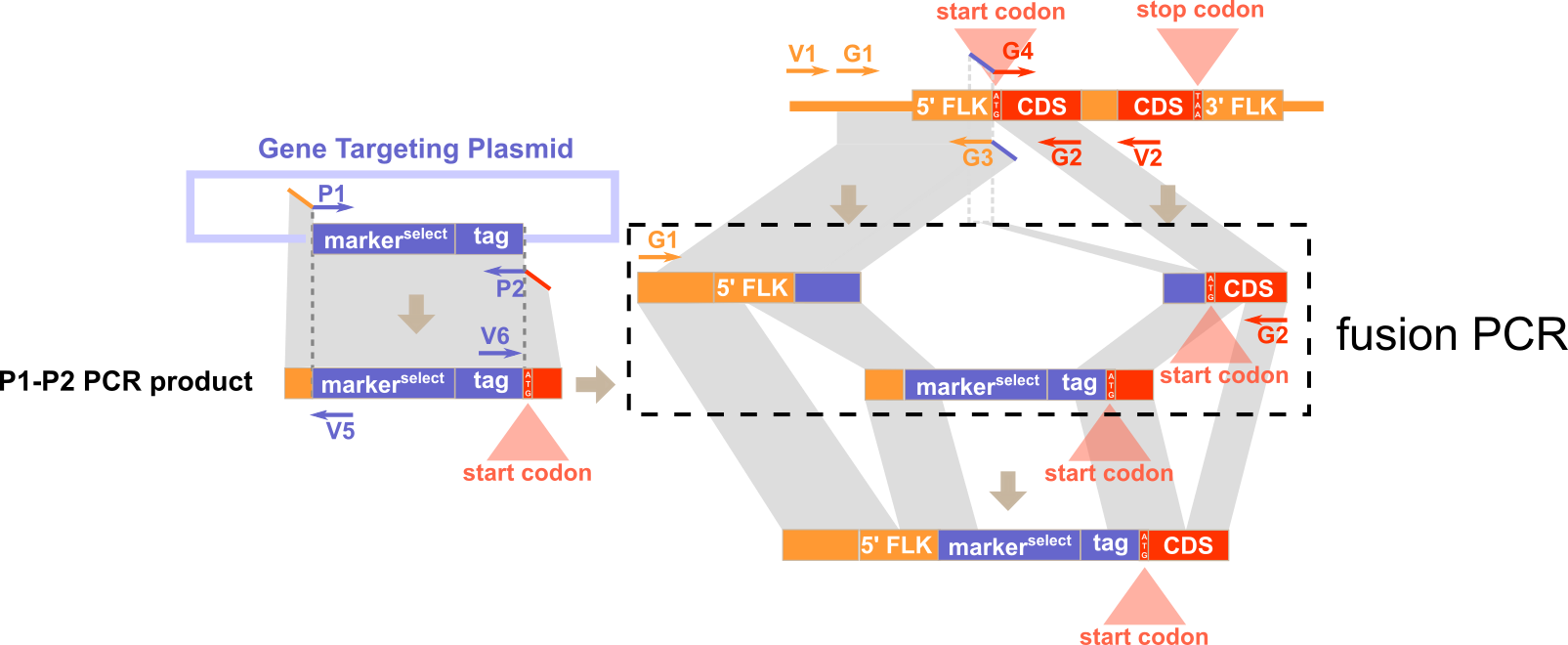
Expected PCR results with verification primers

Note: G3 is located immediately upstream of the start codon and G4 starts from the start codon.
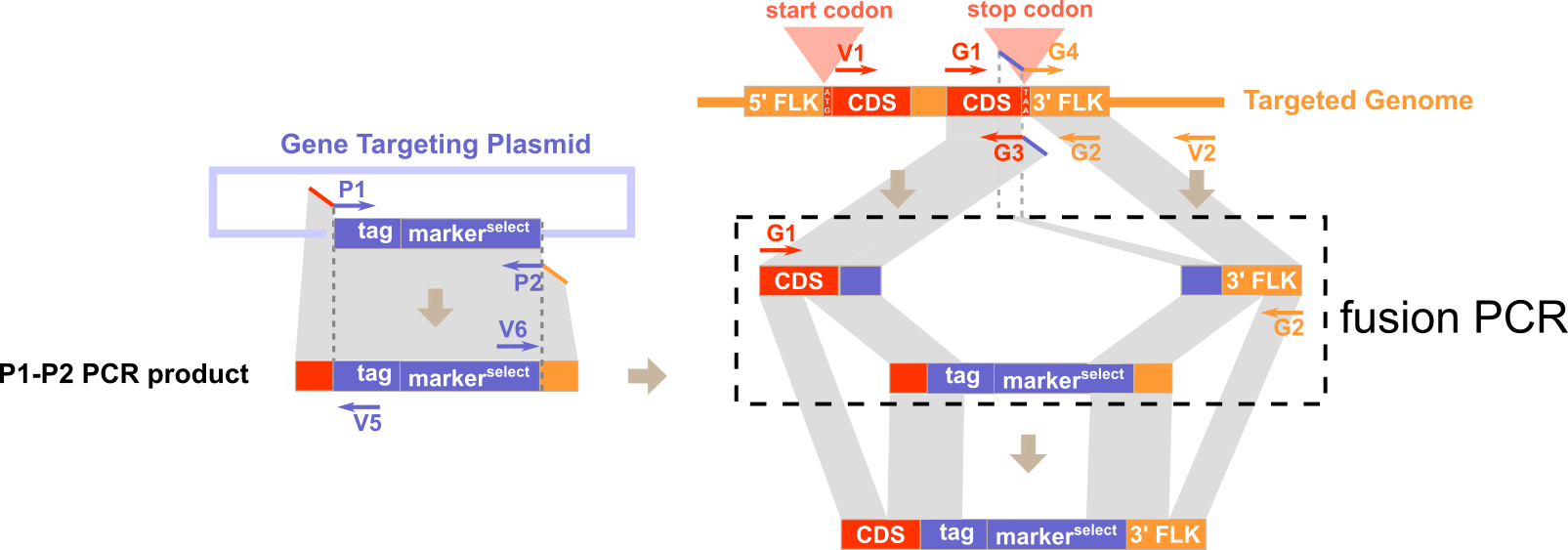
1) P1 and P2 for amplifying the inserted sequences (e.g. selection cassette) from the plasmid while introducing flanking sequence of target gene at the same time.
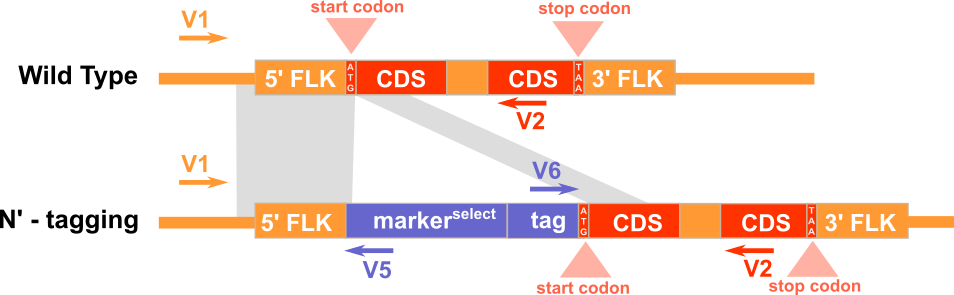
1) V1 and V3 for amplifying the 5'-junction of the WT genomic region before gene targeting. 2) V4 and V2 for amplifying the 3'-junction of the WT genomic region before gene targeting. 3) V1 and V5 for amplifying the 5'-junction the recombinant genomic region after gene targeting. 4) V6 and V2 for amplifying the 3'-junction the recombinant genomic region after gene targeting.
Experimental Design
Expected PCR results with verification primers


Note: Portion of P1 that is homologous to flanking sequence of target gene does not contain start codon and portion of P2 that is homologous to flanking sequence of target gene does not contain stop codon.
Experimental Design
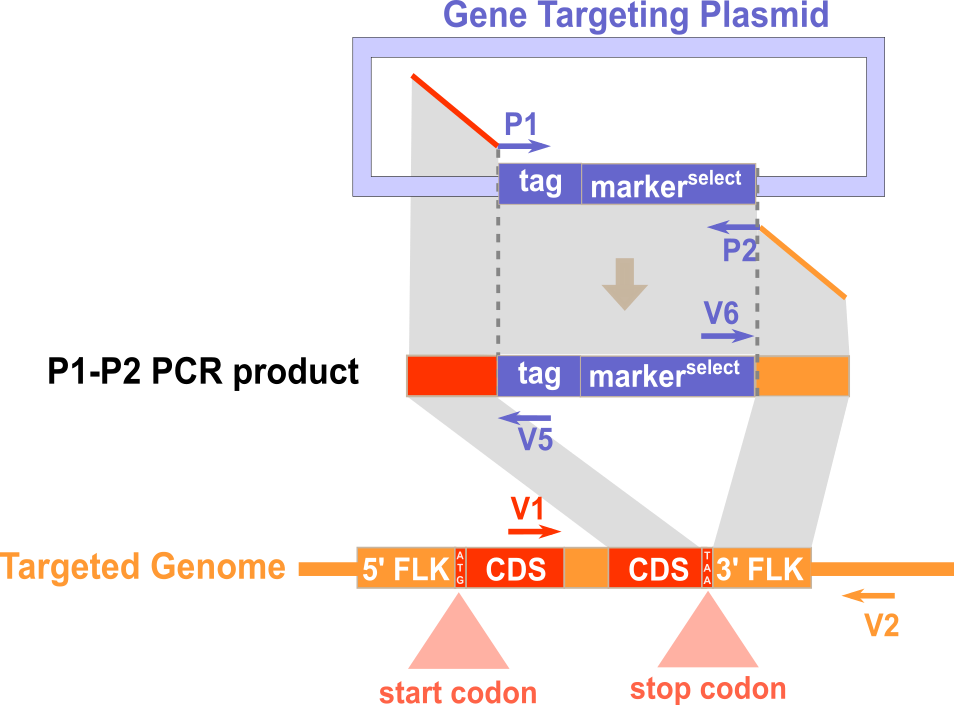
Expected PCR results with verification primers


Note: Neither portion of P1 nor portion of P2 that is homologous to flanking sequence of target gene contains stop codon.
Experimental Design
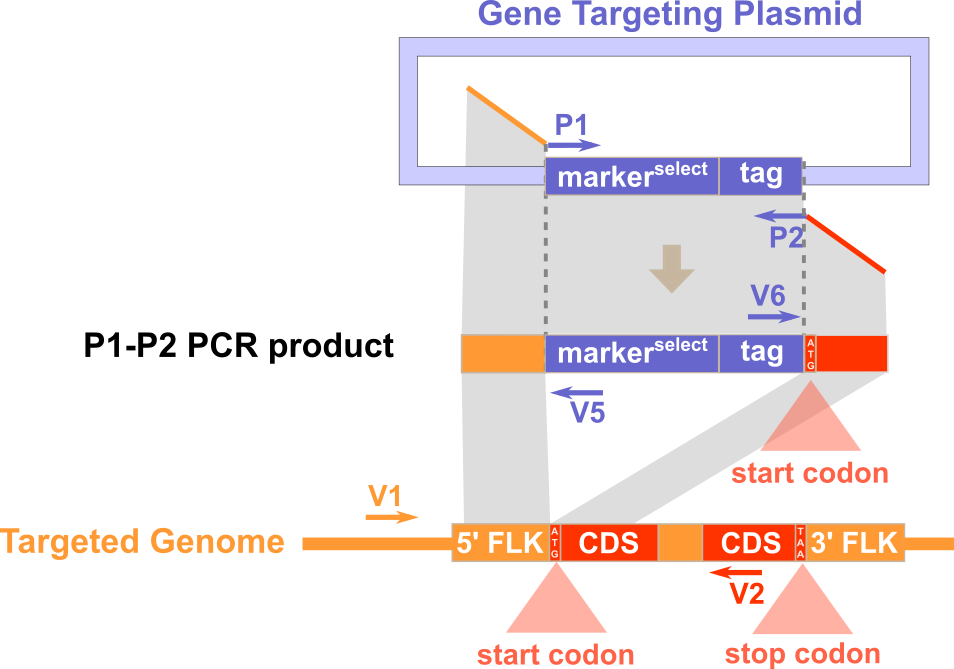
Expected PCR results with verification primers


Note: Portion of P1 that is homologous to flanking sequence of target gene does not contain start codon and portion of P2 that is homologous to flanking sequence of target gene contains start codon.
Given that those widely used gene targeting plasmids (e.g. pFA6) usually have pre-designed common primer seqeunces enclosing the sequences to be inserted (e.g. selection cassette). In this case, GetPrimers only need to know the common primer sequences and their enclosed sequences can be denoted by "NNNNNN".
| Name | Sequence | Type |
|---|---|---|
| pFA6_knockout.fa | CGGATCCCCGGGTTAATTAANNNNNNGTTTAAACGAGCTCGAATTC | knockout |
| pYES_knockout.fa | AGCTTTTCAATTCAATTCATNNNNNNGGAAGAGTATGAGTATTCAA | knockout |
| pCSN44_hph_knockout.fa | GATTTCAGTAACGTTAAGTGGATNNNNNNCTCCTTCAATATCATCTTCTGTC | knockout |
| pBS10_hyg_knockout.fa | CGCCAGATCTGTTTAGCTTGNNNNNNATCCAGTGTCGAAAACGAGC | knockout |
| pKOV21_HygR_knockout.fa | CTTGGCTGGAGCTAGTGGAGGTNNNNNNGAATAGAGTAGATGCCGACCGGG | knockout |
| pUG6_kanMx_knockout.fa | GCTGCAGGTCGACAACCCNNNNNNCTAGTGGCCTATGCGGCC | knockout |
| pUG72_KIURA3_knockout.fa | GCTGCAGGTCGACAACCCNNNNNNCTAGTGGCCTATGCGGCC | knockout |
| pFA6_ctag.fa | CGGATCCCCGGGTTAATTAANNNNNNGTTTAAACGAGCTCGAATTC | C' tagging |
| pFA6_GST_ntag.fa | GAATTCGAGCTCGTTTAAACNNNNNNAATCGGATCTGGTTCCGCGT | N' tagging |
| pFA6_3HA_ntag.fa | GAATTCGAGCTCGTTTAAACNNNNNNCAGATTACGCTGCTCAGTGC | N' tagging |
| pFA6_GFP_ntag.fa | GAATTCGAGCTCGTTTAAACNNNNNNGCATGGATGAACTATACAAA | N' tagging |
| User defined | User defined | User defined |
| Tag | Description |
|---|---|
| ID | Gene ID in the GFF3 file. |
| Gene_Name | Gene name in the GFF3 file. |
| Dbxref_ID | Dbxref ID in the GFF3 file. |
| Strand | The coding direction of the targeted gene: +(forward strand) or - (reverse strand). |
| Chr | The name of the chromosome or scaffold in the GFF3 file. |
| CDS_Start | The starting coordinate of the targeted gene's coding region. |
| CDS_End | The ending coordinate of the targeted gene's coding region stop codon. |
| Gene_Start | The starting coordinate of the targeted gene. |
| Gene_End | The ending coordinate of the targeted gene. |
| Primer_ID | The IDs of designed primers. Note that some initial primer candidates might have been filtered out because of the poor matches with the targeted genome, so the final ID might have been discontinuously numbered. |
| G* | Gene targeting primers, where '*' can be 1, 2, 3 or 4. These primers are designed based on the targeted genome seqeunce. |
| P* | Plasmid primers, where '*' can be 1 or 2. These primers are designed based on the plasmid sequence. |
| V* | Verification primers, where '*' can be 1, 2, 3, 4, 5 or 6. The primer V1, V2, V3 and V4 are designed based on the targeted genome sequence whereas V5 and V6 are designed based on the plasmid sequence. |
| x_Loc | The genomic coordinates of primer X, where X can be one of the following: G1, G2, G3, G4, V1, V2, V3, V4, V5 and V6. |
| x_Tm | The melting temperature (℃) of primer X, where X can be one of the following: G1, G2, G3, G4, V1, V2, V3, V4, V5 and V6. The Tm of G3, G4, P1 and P2 are estimated based on the formulae specified via the corresponding option (see Manual for details), whereas the those of others are estimated by Primer3. |
| x_y_Products_Counts | The number of all possible PCR products expected from the primer pair X and Y based on genome-wide BLAST. |
| x_y_Blast_Len | The estimated sizes of all possible PCR products from the primer pair X and Y based on genome-wide BLAST. Different sizes are seperated by semicolon. |
| x_y_Expect_Len | The estimated size of the true on-target PCR product from the primer pair X and Y. |
| x_y_Dimer | Whether the primer pair X and Y is likely to form a dimer: "1" for yes and "0" for no. |
| x_y_Penalty | The Primer3 penalty of the primer pair X and Y. |
| P*_Blast_Match | The number of noticeable plasmid primer match (alignment identity > 80% and alignment coverage > 80%) with the targeted genome (which might cause off-target recombination), where '*' can be 1 or 2. |
| Tier_Ranking | The tier ranking of the primer set based on various primer quality check criteria (e.g. off-target possibility, primer3 penalty, etc) with Tier 1 ranked at the top. |
| Warnings | Warning messages based on the primer quality check of Primer3. |
Presented by EvomicsLab@SYSUCC yuejiaxing[at]gmail[dot]com